[ad_1]
Graph neural networks and transformers are redefining what is possible in computational medicine by taking advantage of contextual information and large unannotated multimodal datasets.
A graph neural network is a type of machine learning algorithm that uses graph structures to encode spatial relationships between objects. In the context of pathology and radiology, these algorithms are being used to take advantage of spatial context for a variety of applications, including image segmentation, disease classification, and tissue analysis.
One of the main challenges in medical imaging is to accurately identify and segment objects of interest such as tumors or organs. Traditional machine learning algorithms often struggle with this task due to the complex spatial relationships within medical images. Graph neural networks, on the other hand, are able to explicitly encode these relationships, allowing for more accurate object segmentation.
For example, a graph neural network can be used to segment an image of the brain into different types of tissue, such as gray matter and white matter. The algorithm will first build a graph structure, where each node represents a pixel in the image and each edge represents a spatial relationship between two pixels. The algorithm would then use this graph structure to learn the spatial relationships between pixels and identify different tissue types.
In addition to image segmentation, graph neural networks are also being used for disease classification. For example, a graph neural network can be trained to recognize whether an image of a lung contains a tumor. The algorithms would build a graph structure that encodes the spatial relationships between different regions of the lung, and then use this structure to learn the spatial patterns associated with healthy and diseased tissue.
Another application of graph neural networks is tissue analysis in pathology and radiology. It involves using spatial relationships within an image to understand the underlying biology of a tissue sample. For example, a graph neural network can be used to analyze the spatial organization of cells within a tissue sample, allowing for more accurate diagnosis and prognosis of diseases.
Overall, graph neural networks are taking advantage of spatial context in pathology and radiology to improve upon traditional machine learning algorithms. By explicitly encoding the spatial relationships within images, these algorithms are able to improve the accuracy of tasks such as image segmentation, disease classification, and tissue analysis. This has the potential to greatly improve the ability of medical professionals to diagnose and treat diseases and ultimately improve patient outcomes.
The first six paragraphs of the main text were not written by the editors Nature Biomedical Engineering, They were quickly generated by ChatGPT (Generative Pre-Training Chat) – a prototype general-purpose chatbot based on the GTP3.5 Transformer model.1Encoder-decoder architecture and a large language model with approximately 200 billion parameters – by inputting the prompt “Discuss, in approximately 600 words, how graph neural networks are taking advantage of spatial context for use in pathology and radiology.”
Transformer models are powerful. ChatGPT is trained on Internet corpora available up to 2021, using self-supervised learning tasks, such as masked-language modeling, and learning the meaning of words through contextual clues through a self-attention mechanism2 (Methods that augment parts of the input data so as to improve their overall representation; for example, the self-attention algorithm considers the context of a word in the input sequence for clues that give the word a numerical representation.) which preserves the relationship between the words in the sentence). Before applying self-attention, transformers map tokenized text to embeddings (continuous low-dimensional vector representations of discrete variables) and use positional encoding for word order. Cross-attention (that is, computing an attention score for one type of data via information from another type of data, such as other embeddings) and multi-head attention (or multiple attention mechanisms running in parallel) in decoding the output Are included.
Therefore, transformers learn the meaning of words and the structure of language through contextual cues. By repeatedly predicting the next word based on an input query, they can create new text, summarize stories, reorder scrambled text, identify topics in discussion, code can write and do a myriad of other things3, all in full prose. Because transformers can learn representations of arbitrary input data, they are also used to build models that generate images from textual signals. The quality of the output of any such model largely depends on the quality of the data with which they have been trained. Spurious joins in the data can mislead the model; They can be confidently treated as providing plausible but inaccurate information, and can easily be corrected to existing datasets (Figure 1).
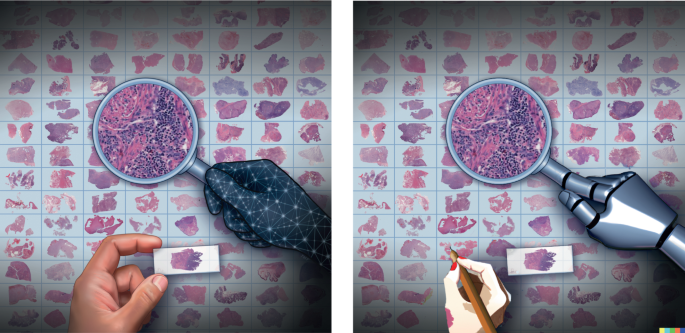
Katie Yost and OpenAI.
The image on the right corresponds to an output from OpenAI’s Transformer model DALL-E 2 when it is asked to sequentially modify the hands on the image on the left according to the cues of “a real female hand”. and “An arm from an AI robot.”
Because one of the main advantages of transformers is that they do not require explicit annotation in the data with which they are trained, they can also be used to build models for classifying images without explicit supervision. As shown by Pranav Rajpurkar and colleagues (E. Tiu et al. nuts. Biomed. England, https://doi.org/10.1038/s41551-022-00936-9; 2022) to detect the presence of distortions in unlabeled chest X-ray images. In this case, the model uses a text transformer and a vision transformer to learn pathology features from the raw radiology report associated with each X-ray image using contrast learning (a method for predicting whether pairs of samples are related or unrelated). ) can be used. , Therefore, radiology reports serve as a natural source of contextual clues for the model to learn pathology features in X-ray images.
As ChatGPT’s outputs relayed above suggest, spatial context can improve the performance of machine-learning tasks in pathology. Graph representations – that is, sets of interconnected nodes and edges – are particularly well suited for any system that can be mapped as a network, such as networks of cells or tissue patches in medical images, and for medical codes. Concepts Records in Networks and Electronic Health, as discussed in a perspective article by Marinka Zitnik and colleagues (MM Lee et al. nuts. Biomed. England, https://doi.org/10.1038/s41551-022-00942-x; 2022). this issue Nature Biomedical Engineering Also included are three examples of the benefits of applying graph neural networks to tasks in pathology.
In an article, James Xu, Aaron Meyer and Alexandro Trevino show (Z Wu et al. nuts. Biomed. England, https://doi.org/10.1038/s41551-022-00951-w; 2022) that spatial protein profiles obtained through multiplex immunofluorescence can be used to train a graph neural network to model the tumor microenvironment in tissue samples from patients with head and neck and colorectal cancer to infer spatial motifs. linked to cancer recurrence. And with the survival of the patient after treatment. In another article, Sunghoon Kwon and colleagues showed (Y Lee et al. nuts. Biomed. England, https://doi.org/10.1038/s41551-022-00923-0; 2022) that, in whole-slide images of tumors, graph deep learning can be used to derive interpretable relevant histopathological features in the tumor microenvironment that are predictive of patients’ prognosis. As reported by Faisal Mahmood and co-authors in an associated News & Views (G. Joum et al. nuts. Biomed. England, https://doi.org/10.1038/s41551-022-00924-z; 2022), context-aware graph neural networks can be thought of as sitting between models that do not have access to any context and models (such as vision transformers) that have access to full spatial context, and those with inductive biases or assumptions. include a degree of variance that allows the model to generalize. In another research article in this issue, Mahmoud and colleagues show another application of self-supervised learning: searching and retrieving gigapixel full-slide images (Figure 1) at a speed independent of the size of the repository (C. Chen et al). nuts. Biomed. England, https://doi.org/10.1038/s41551-022-00929-8; 2022). To discover tissue patches, instead of querying against each slide in the dataset, a variational autoencoder (a probabilistic generative model that learns latent representations of data) is selected to select patches from each slide as a set of codes. trained to represent. That the patch with the highest probability of matching the query can be retrieved by taking advantage of uncertainty-based ranking and tree data structure for speed efficiency and scalability.
For applications in medicine and health, openly releasing pre-trained models may facilitate the shift from model building to model deployment in health care settings, as argued by Joseph Wu and colleagues in a review article is (A Zhang et al. nuts. Biomed. England, https://doi.org/10.1038/s41551-022-00898-y; 2022) in this issue. This is because Transformers, graph representation learning, and many other self-supervised deep-learning models require large unlabeled datasets for pre-training, which improves the performance of downstream tasks. In fact, Ryan Krishnan, Pranav Rajpurkar and Eric Topol have published a review article (R. Krishnan et al. nuts. Biomed. England, https://doi.org/10.1038/s41551-022-00914-1; 2022), “Self-supervised learning leveraging multimodal data will enable the construction of high-performance models that better ‘understand’ the underlying physiology.” Trained Chatbot seems to agree with this, adding that “quality context is critical for good performance of ChatGPT”.
[ad_2]
Source link

